Vision
Our goal is to create a unified and dynamic resource to enhance basement membrane research.
Our vision for basement membraneBASE is to create an atlas of basement membrane composition during development, adult life, disease and across animal species formed by connecting basement membrane gene networks with protein localisation data.
We will also share protocols for studying basement membranes.
Background
Basement membranes are supramolecular assemblies that form membrane-like matrices underlying all continuous sheets of cells such as epithelia and endothelia.
Basement membranes are built on two self-assembling networks – laminin (composed of α, β and γ chains), and type IV collagen (composed of three α chains). The covalently cross-linked type IV collagen network provides tissues with mechanical support, while laminin anchors basement membranes to tissues through integrin and dystroglycan cell surface receptors.
These networks interact with the other core basement membrane components nidogen, agrin, and perlecan, as well as with various matricellular proteins such as fibulins, spondin, and type XVIII collagen, which provide basement membranes distinct biophysical and biochemical properties.
In addition to mechanical support, basement membranes have barrier and filtration functions (e.g., blood brain barrier, kidney glomerulus), direct epithelial polarity, and harbour growth factors and proteases that mediate cellular differentiation, survival, and migration.
Genetic disruption of basement membranes in mice, Caenorhabditis elegans and Drosophila results in embryonic lethality due to their essential roles. In humans, variants in basement membrane genes underlie many developmental and adult onset disorders affecting numerous tissues (e.g., eye, brain, kidney, muscle, vasculature, skin).
Further, defects in basement membrane component expression and turnover is a key pathogenic aspect of diseases such as cancer, fibrosis, and diabetes, and is an important factor in aging.
Understanding basement membrane composition and the mechanisms that regulate basement membrane formation, growth, and turnover are of fundamental importance to human health.
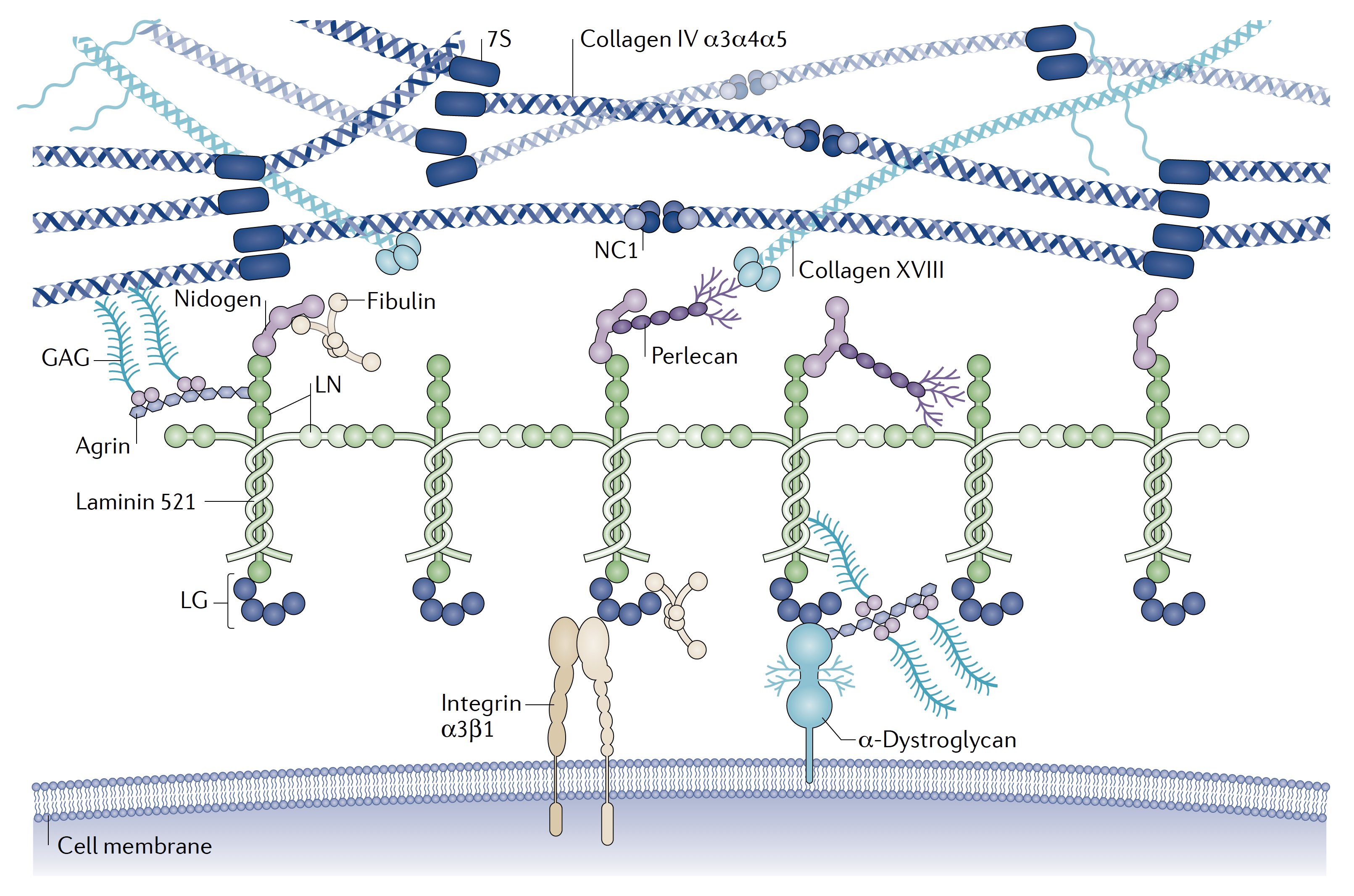
The core components of basement membranes. Adapted from: Complexities of the glomerular basement membrane, Nature Reviews Nephrology PMID: 32839582
Gene network
We generated a comprehensive, integrated network of 222 human genes encoding BM zone proteins: 160 of these encode BM components and 62 are cell surface interactors, such as BM receptors.
Summary of gene network data
- 222 BM zone human genes
Download this data as an Excel spreadsheet (53.1KB) - 184 genes with confirmed evidence of protein localisation to the BM zone (from protein immunolocalisation studies)
- 160 components with confirmed evidence of protein localisation to the human BM zone
- 38 genes predicted to be in the BM zone based on protein interaction data or BM protein-cleaving protease activity.
- 222 genes: 67 glycoproteins, 24 collagens, 40 regulators, 15 proteoglycans, 13 ECM-affiliated proteins, 6 secreted factors and 58 other molecules, including 37 well-characterised BM receptors.
- Database links to localisation evidence
- Main publication
Future work on this network will include new component discovery, functional animal modelling and interrogation of patient genomes.
Localisation evidence
You can access localisation data for a number of organisms in our database.
To search our database, click below.
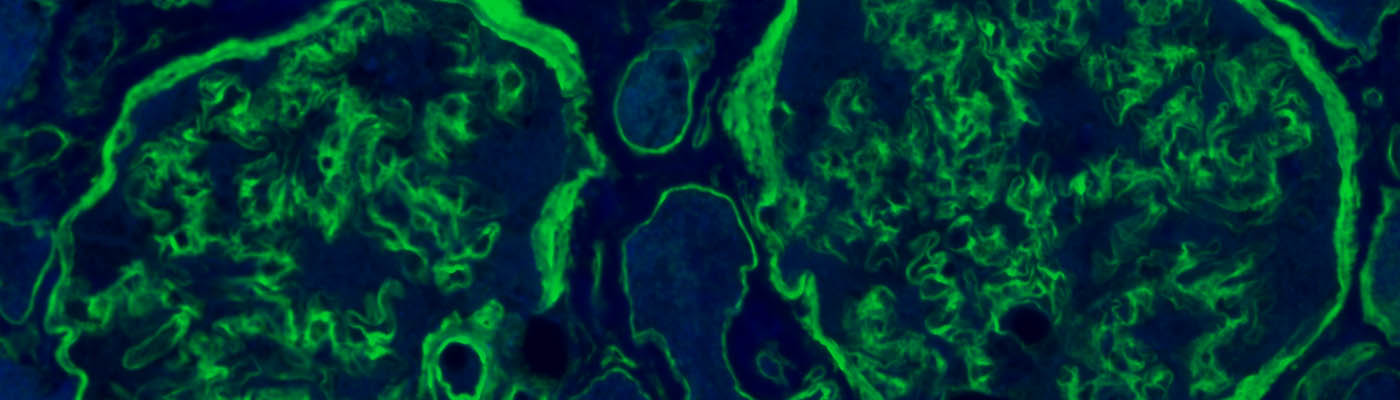
Collagen IV immunofluorescence identifying the basement membrane and mesangial matrix in human kidney tissue.
Major basement membrane components
Information on a number of basement membrane components.
Find details of major and well-studied basement membrane components and their links to human disease.
Experimental systems
There are a number of experimental systems that are suitable for studying basement membrane biology.
We highlight the range of these systems and their relative strengths.
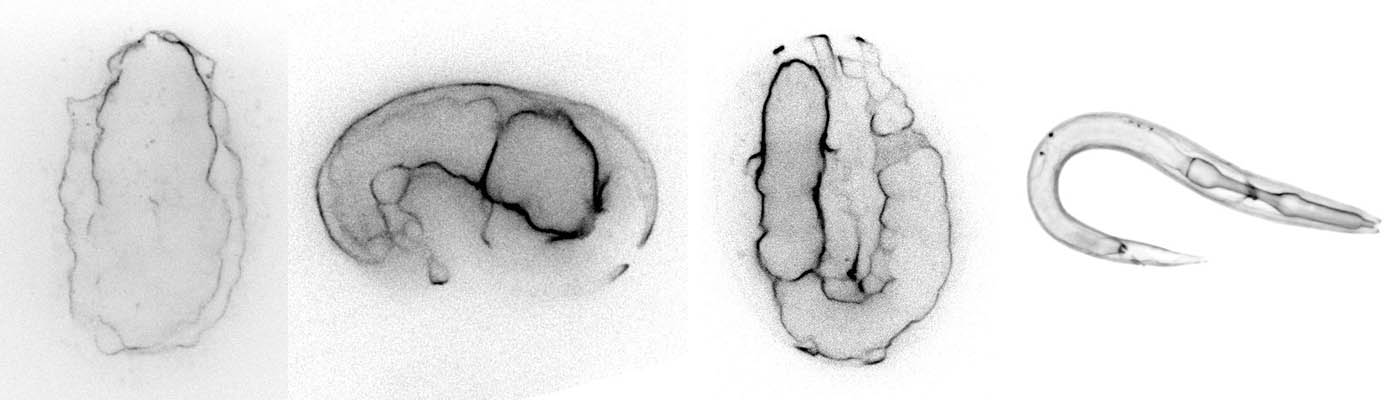
The model organisms C. elegans during embryonic development.
Reagents and protocols
Access reagent and protocol information relating to our data.
![]() Antibody lists (Gene list, Table 1, List 1)
Antibody lists (Gene list, Table 1, List 1)
C. elegans
Contributors

Collaborators at Duke University and Manchester University during the creation of basementmembraneBASE.
The University of Manchester
- Bernard Davenport
- Maryline Fresquet
- Jack Ingham
- Nikki-Maria Koudis
- Craig Lawless
- Anna Li
- Mychel Morais
- Richard Naylor
- Pinyuan Tian
Duke University
- Qiuyi Chi
- Eric Hastie
- Ranjay Jayadev
- William Ramos-Lewis
- Sandy Srinivasan
IT team
- Anna Fildes
- Andrew Jerrison
- Veselin Karaganev
- Helen Robinson
- Joshua Woodcock
Contact us
Please get in touch if you have any questions about basement membraneBASE.
Rachel Lennon
The University of Manchester, UK
Email: Rachel.Lennon@manchester.ac.uk
David Sherwood
Duke University, USA
Email: david.sherwood@duke.edu
Useful links
- Lennon lab (Wellcome Trust Centre for Cell Matrix Research)
- Lennon Lab website
- Sherwood lab (Duke University)
- Matrisome project (University of Illinois Chicago)
- WormBase
- Expression Atlas (EMBL-EBI)
- Gene Ontology Resource
- The Matrixome Project